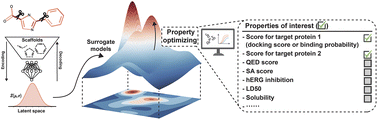
Designing molecules with multiple properties of interest is a fundamental challenge in drug development. To tackle this, we have developed ScafVAE, an innovative scaffold-aware variational autoencoder designed for the in silico graph-based generation of multi-objective drug candidates. By integrating our proposed bond scaffold-based generation with perplexity-inspired fragmentation, we expand the accessible chemical space of the conventional fragment-based approach while preserving its high chemical validity. ScafVAE was pre-trained on a large dataset of molecules and further augmented through contrastive learning and molecular fingerprint reconstruction, resulting in high accuracy in predicting various computationally and experimentally measured molecular properties. Only a few of its parameters are task-specific, facilitating easy adaptation to new desired properties. ScafVAE was employed to generate dual-target drug candidates against drug resistance in cancer therapy, considering four distinct resistance mechanisms, with or without additional properties such as drug-likeness or toxicity. The generated molecules exhibited strong binding strength to target proteins using molecular docking or experimentally measured affinity while maintaining optimized extra properties. Further molecular dynamics simulations confirmed the stable binding interactions between the generated molecules and target proteins. These findings position ScafVAE as a promising alternative to conventional generation approaches.