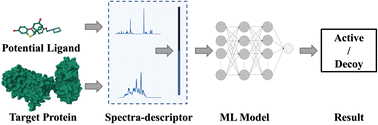
Machine learning models have emerged as powerful tools for drug discovery of lead compounds. Nevertheless, despite notable advances in model architectures, research on more reliable and physicochemical-based descriptors for molecules and proteins remains limited. To address this gap, we introduce the Fragment Integral Spectrum Descriptor (FISD), aimed at utilizing the spatial configuration and electronic structure information of molecules and proteins, as a novel physicochemical descriptor for virtual screening models. Validation demonstrates that the combination of FISD and a classical neural network model achieves performance comparable to that of complex models paired with conventional structural descriptors. Furthermore, we successfully predict and screen potential binding ligands for two given protein targets, showcasing the broad applicability and practicality of FISD. This research enriches the molecular and protein representation strategies of machine learning and accelerates the process of drug discovery.