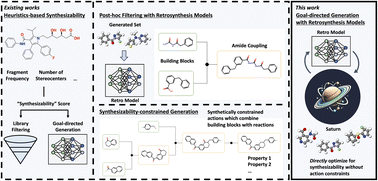
Synthesizability in generative molecular design remains a pressing challenge. Existing methods to assess synthesizability include heuristics-based metrics or retrosynthesis models which predict a synthetic pathway. By contrast, an explicit approach anchors generation with “synthetically-feasible” chemical transformations, such that all generated molecules already have a predicted synthetic pathway. To date, retrosynthesis models have been mostly used as a post hoc filtering tool as their inference cost remains prohibitive to use directly in an optimization loop. In this work, we show that with a sufficiently sample-efficient generative model, it is straightforward to directly optimize for synthesizability using retrosynthesis models in goal-directed generation. Under a heavily-constrained computational budget, our model can generate molecules satisfying multi-parameter drug discovery optimization tasks while being synthesizable, as deemed by retrosynthesis models. We reaffirm previous findings that common synthesizability heuristics (formulated based on known bio-active molecules) can be well correlated with retrosynthesis models’ solvability, such that optimizing for the latter may not be an optimal allocation of computational resources. However, going further, we show that moving to other classes of molecules, such as functional materials, current heuristics’ correlations diminish, such that there is an advantage to incorporating retrosynthesis models directly in the optimization loop. Finally, we demonstrate that over-reliance on synthesizability heuristics can overlook promising molecules. The codebase is available at https://github.com/schwallergroup/saturn.